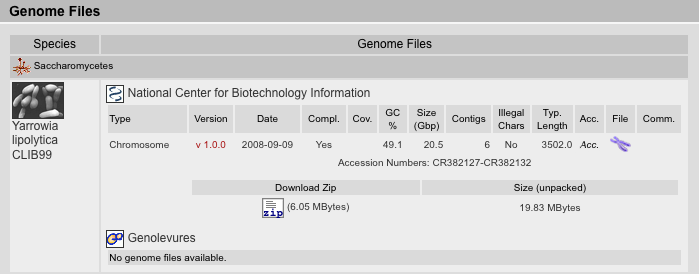
The Genome Files Result View lists information corresponding to genome assembly files. The files are sorted by species followed by sequencing centers. The type of the file is inserted manually and can be one of the following:
| Chromosome | Every fasta-entry in this file is a chromosome. There are a few exceptions where chromosomes are split in two or more fasta-entries. |
| Uchromosome | These files contain contigs/supercontigs which could not be mapped to any (unknown chr.) or anchored (random chr.) to a certain chromosome. |
| Supercontigs | Every fasta-entry represents a supercontig which consist of sorted contigs separated by estimated or fixed numbers of "N" bases. |
| Contigs | Contigs (from "contiguous sequence") are the smallest pieces of an assembly and consist of overlapping sequence reads. |
| Ureads | These files contain the unplaced reads, reads that could not be assembled to contigs. These files are especially important for low-coverage genomes that in most cases end up with very short contigs. In these cases, small proteins or some exons can be reconstructed from the ureads-files. |
| Apicoplast | These files contain the apicoplast DNA. The apicoplast is a relict, non-photosynthetic plastid found in Apicomplexa. It is proposed that it evolved via secondary endosymbiosis. The apicoplast is surrounded by four membranes within the outermost part of the endomembrane system. |
| Chloroplast | These files contain the chloroplast DNA. Chloroplasts are organelles found in plant cells and eukaryotic algae that conduct photosynthesis. |
| Kinetoplast | These files contain the kinetoplast DNA. A Kinetoplast is a disk-shaped mass of circular DNAs inside a large mitochondrion that contains many copies of the mitochondrial genome. Kinetoplasts are only found in protozoa of the class kinetoplastea. Kinetoplasts are usually adjacent to the organisms' flagellar basal body leading to the thought that they are tightly bound to the cytoskeleton. |
| Mito | These files contain the mitochondrial DNA. Mitochondria are membrane-enclosed organelles found in most eukaryotic cells. |
In addition, there are other rarely used file types: Ultracontigs (very long supercontigs, but not mapped to chromosomes yet), Usupercontigs (contigs that could not be ordered to supercontigs).

Where available, we provide the version of the assembly as well as the release date of the data. In general, we have taken the versions and release dates as given by the sequencing centers. If those are not provides, we have taken the dates on which the files were saved in the ftp-directories. For NCBI-assembly data, we have taken the dates on which the data has been submitted to NCBI. Note: Version numbers do not correlate between sequencing centers and NCBI! Assembly version 6.0 at a sequencing center might correspond to version 1.0 at NCBI because it was the first version submitted.
The completeness is the same as given in the projects view, and is a rough estimate of the completeness and quality of the data and assembly. In general, assemblies with coverages below 4 are regarded as incomplete.
The genome coverage of the assembled sequence data is given if it is provided by the sequencing centers.
The GC content, the size in Giga-base-pairs, the number of fasta-entries ("contigs"), the occurrence of illegal characters in the sequences (not beeing g/G, a/A, t/T, c/C, or n/N), and the typical length of the fasta-entries were calculated from the fasta files.
For genome assemblies available from NCBI, the accession numbers can be shown by clicking on "Acc." and the assemblies are provided as zipped fasta files.
For some assemblies, comments are available that provide further background information about differences to earlier assemblies, problems in the assembly process, and others.